Hello,
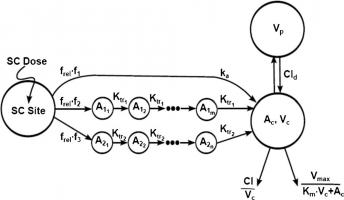
I have data of ER injection which would release the drug over longer period of time.I have modelled the data using transit compartment modelling.I am attaching phx prj file in which i have tried 2 different approaches (2cmt_2 depot_estKel_add & 2cmt_3depot_estKel_add)
- 2cmt_2 depot_estKel_add: In this approach i have divided the dose into 2 different fractions ie 1 fraction directly reaches the central cmt and another fraction reaches through transit comp and then reaches the central comp. My data fitting using the model seems to be ok.I need help in further optmising this model
- 2cmt_3depot_estKel_add: In this approach i have divided the dose into 3 different fractions ie 1 fraction directly reaches the central cmt and another 2 fraction's reaches through 2 transit comp's and then reaches the central comp.I have fitted the data to this model but overall the fitting remains the same as that of earlier approach. I need help here in optimising the model and whether my model is correctly written based on my hypothesis stated above.
I need support from people to optmise the models further, your response will be of great help
Regards
HK