Hi Sara,
I did not check the code in much detail, just used your formulas to do:
1. define dissolution rate outside of the deriv statement
2. limit the drate to 0-+Inf
3. added bioavailability (F_p) to the absorption
4. defined the static value Aadose as total dose amount
Here is the code:
test()
{
# GI compartment
deriv(Aa = - drate * F_p * Aadose)
# Central compartment
deriv(A1_p = drate *F_p * Aa - (Cl_p * C_p)- (Q_p * (C_p - C2_p)) - (VMax_p_m * C_p / (C_p + Km_p_m)))
# Peripheral compartment
deriv(A2_p = (Q_p * (C_p - C2_p)))
# Define Weibull for dissolution rate
drate=max(0,((Fmax*(Beta*scB)*(t^((Beta*scB)-1))*exp(-(t^((Beta*scB)))/(Alpha*scA)))/(Alpha*scA)))
# Define Dosepoint
dosepoint(Aa, idosevar=Aadose)
# Concentrations definition
C_p = A1_p / V_p
C2_p = A2_p / V2_p
# Model erro
error(CEps_p = 0.1)
observe(Parent = C_p * exp(CEps_p))
# Initial estimates
# Dissolution (Weibull)
fixef(Fmax(freeze) = c(, 1, ))
fixef(Alpha(freeze) = c(, 7, ))
fixef(Beta(freeze) = c(, 1.5, ))
fixef(scA = c(, 80, ))
fixef(scB = c(, 2, ))
# Central compartment
fixef(V_p = c(0, 88, 400))
fixef(Cl_p = c(0, 28, 60))
fixef(F_p = c(0, 0.05, 0.4))
# Peripheral compartment
fixef(V2_p = c(0, 181, 3000))
fixef(Q_p = c(0, 22, 300))
# Metabolite conversion
fixef(VMax_p_m = c(0, 329, 6000))
fixef(Km_p_m = c(0, 700, 5000))
}
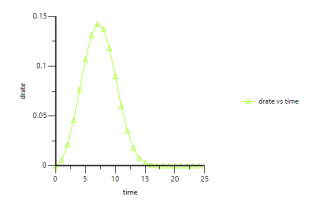
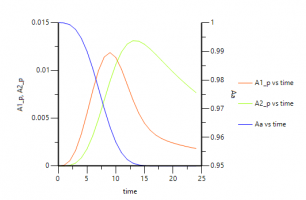
An here are some plots:
Let me know if this does not work for you.
Bernd