Hello and Happy New Year!
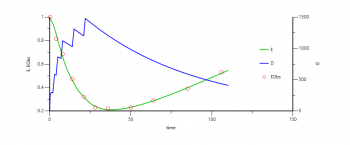
I am trying to write a model for describing Dose-Response-Time data of oligonucleotides using PML. I am using the data from Tsimikas et al. 2015 Lancet 386, 1472-1483 and the model described in Jansson-Lofmark and Gennemark 2018 Nucleic Acid Therapeutics 28 (6), 319-325
Because I am learning how to use PML using the PML school videos (thank you so much for them!), I would like to make sure that I am doing the right thing. Hence, I have attached the raw data and the model code.
 Modeling Dose-Response-Time data.txt 1.14KB
127 downloads
Modeling Dose-Response-Time data.txt 1.14KB
127 downloads
It would be great to know if everything is correct and if I am on the right path.
Many thanks
Annalisa