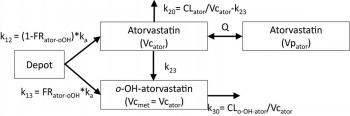
I'm developing a model in which the parent drug is metabolized to an active metabolite and a portion of the oral dose is metabolized in the GI to the active metabolite. I am trying to model the amount that is metabolized from the dose in addition to remaining amount goes to the central compartment of the parent drug.
I found this model created by one member who tried to build the structural model using graphical model, however, he did not account for fraction of the dose went to the parent and metabolite compartments.
Can I get help on how to model the fraction formed?
I am attaching the model and a schematic description of a model that shows the structural model details.