Toxicokinetics, NCA: my plasma data are from sparse sampling in rats after subcut. dosing, time points are 0.083, 0.25, 0.5, 1, 4, 8 h after dose, dosing interval 24h. I would like to predict C24 from lambda_z (calculated with 1,4, 8h) to use the predicted C(24) as C(0) in steady state (day 14). How can I predict C(24)? Thank you for answering.

NCA, Tau=24h, prediction of C(24) if no samples were taken at 24h
#2

Posted 13 June 2016 - 02:29 PM
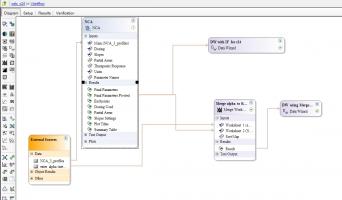
Hi Dieter, please look at the attached project, it's not very elegant but will get you what you need I think. there's probably a better way of doing it if I think a bit more.
1) switch on intermediate calculations in NCA.
2) after executing, look in core output for the line;
Best value for Lambda_z: 0.1428, and intercept: 2.6856
3) use this intercept value in the formula in the subsequent DW, (unfortunately this value is not out put to a worksheet so we can't easily map it in, and therefore if you have several profile you might to code an IF statement to switch the values)
exp((2.6856- (lambda_z*24)))
(where 24 is the time for which you want to predict the conc.)
Simon
Attached Files
#3

Posted 14 June 2016 - 01:34 PM
Dieter sent me a PM to clarify a few things so also posting here for others;
DW is my abbreviation for Data Wizard (see Data Tools), this is very handy to do all the stuff you may have been doing in Excel sheets before since it menas it is all containted in your Phoenix project and your workflow can be easily updated at anytime. You’ll see an expanded example of it in the attached project where I've expanded the dataset to have three profiles.
I’ve done it two ways, one using a DW with IF but just for subject 1 and then the same value for all others, since as you see it gets a bit complicated. The syntax for these functions is all in the manuals and/or in the right hand panel, you can click and drag functions from there.
The other I refined the workflow to be more friendly I think; creating a second work sheet to enter my alpha values into in the data folder. I could then merge them in before having a simpler code in my data wizard as you can see in these two objects. This means there is a much simple manual step in copying the alpha intercepts out of the Core output.
(Incidentally this idea of requesting conc at a certain time is already noted as a possible enhancement;
QC: 3174. NCA – new parameters C(t) like C8, C24 (concentration at a certain time value), interpolated if needed following AUC method)
Simon
PS If you want more tips like this don't forget to try our training courses;
http://www.certaraun...e_id=23&login=1
Attached Files
Edited by Simon Davis, 14 June 2016 - 01:34 PM.
#4

Posted 15 June 2016 - 01:37 PM
Hi Simon and Dieter,
there is a way to get intercept without core output and copy-paste method.
1. Merge Slopes settings output sheet in NCA with profiles
You will get a dataset Subj-Time-conc-Start time - End time
2. Filter with Data Wizard time points that more than End time and less than Start time
3. Log-transform the conc column
4. Send the result to object Linear in WNL5 Classic Modeling, select 502 - Linear regression
5. Sort - Subj, x - time, y - logconc
6. Execute and you'll see Final Parameters Pivoted sheet
7. INT is desired intercept, SLOPE is -lambdaz.
By the way Simon is right, if you want more tips (for example in case with some excluded points in Slopes Selector) try training courses! ![]()
#5

Posted 17 June 2016 - 08:00 AM
@mittyright: thank you
To be honest, I am very surprised that there is no simple straigt forward function to predict C(t), e.g. C(24), within NCA. A prediction of AUC(0-24) is easily possible via "partial areas" If this is possible, than C(24) should also be simpy predictable. Simon wrote me, that this is on a "to do" list.
#6

Posted 17 June 2016 - 08:33 AM
Hi Dieter, any concentration value is of course being predicted 'behind the scenes' for partial areas etc. It is just these have not been exposed as an option to the user and historically there has not been a lot of people asking for it from e.g support cases so it's never been prioritised.
However your point is noted and althoguh there is are several work arounds (actually I may be able to refine Mitty's suggestion when I have some spare time to fully automate it); we will be moving this request up the priority list as recently more users have asked for this; at this time i cannot give a time line though.
Simon.
#7

Posted 25 June 2016 - 05:31 PM
Hi Dieter et al.
we can predict concentrations for any time point from the standard output of NCA. Yes, we need a little workaround. ;-)
Consider these relationships – which are employed by PHX internally:
- AUCINF_pred = AUClast + Clast_pred / Lambda_z
- Clast_pred = C0 * exp(-Lambda_z * Tlast)
We are interested in C0. With a little algebra:
C0 = (AUCINF_pred - AUClast) * Lambda_z / exp(-Lambda_z * Tlast)
Note that C0 is in untransformed (raw) scale. The intercept given in the NCA core output and INT by mittyright’s workaround is ln(C0).
Now we can predict concentrations for any T by:
CT = C0 * exp(-Lambda_z * T)
NCA: Final Parameters Pivoted > Data Wizard > Add Transformation > Transformation Type: Custom
In your case: New Column Name C24, Formula
(AUCINF_pred - AUClast) * Lambda_z / exp(-Lambda_z * Tlast) * exp(-Lambda_z * 24)
Voilà – just one step… ;-)
But: Don’t search for Clast_pred in NCA | Final Parameters. It’s my invention. Luckily we don’t need it because it cancels out anyway.
If you want to see it the procedure needs more steps:
- NCA: Final Parameters Pivoted > Data Wizard > exclude everything except Lambda_z, Tlast, AUCINF_pred (and your sort columns like Subject, Treatment, etc. of course).
- NCA: Summary Table > Data Wizard > exclude everything except Time, Predicted and your sort columns.
- Join both results. ☑ Inner Join (important!).
1st worksheet: Sort for the columns as above + Tlast, source column Lambda_z, AUCINF_pred
2nd worksheet: Sort for the columns as above + Time, source column Predicted
Click on Sort Map, ☑ Use internal Worksheet:
In the top row(s) there should be our usual mappings.
In the last row: Column Worksheet 1 Sorts: Tlast, Worksheet 2 Sorts: Time. - Execute to see the predicted concentrations at Tlast.
Hope that helps.
Prioritising this issue would be nice. C(TAU) is one of the mandatory PK metrics according to the EMA’s MR guideline (11/2014). If the half life is relatively short and there are deviations from scheduled sampling at TAU using the predicted C(TAU) will reduce variability. Helps a lot if the concentration at TAU is missing (like in Dieter’s case). I’m using this approach for years.
- DieterZ likes this
#8

Posted 27 June 2016 - 07:56 AM
Hi Dieter et al.
we can predict concentrations for any time point from the standard output of NCA. Yes, we need a little workaround. ;-)
Consider these relationships – which are employed by PHX internally:
- AUCINF_pred = AUClast + Clast_pred / Lambda_z
- Clast_pred = C0 * exp(-Lambda_z * Tlast)
We are interested in C0. With a little algebra:
C0 = (AUCINF_pred - AUClast) * Lambda_z / exp(-Lambda_z * Tlast)
Note that C0 is in untransformed (raw) scale. The intercept given in the NCA core output and INT by mittyright’s workaround is ln(C0).
Now we can predict concentrations for any T by:
CT = C0 * exp(-Lambda_z * T)
NCA: Final Parameters Pivoted > Data Wizard > Add Transformation > Transformation Type: Custom
In your case: New Column Name C24, Formula
(AUCINF_pred - AUClast) * Lambda_z / exp(-Lambda_z * Tlast) * exp(-Lambda_z * 24)
Voilà – just one step… ;-)
But: Don’t search for Clast_pred in NCA | Final Parameters. It’s my invention. Luckily we don’t need it because it cancels out anyway.
If you want to see it the procedure needs more steps:
- NCA: Final Parameters Pivoted > Data Wizard > exclude everything except Lambda_z, Tlast, AUCINF_pred (and your sort columns like Subject, Treatment, etc. of course).
- NCA: Summary Table > Data Wizard > exclude everything except Time, Predicted and your sort columns.
- Join both results. ☑ Inner Join (important!).
1st worksheet: Sort for the columns as above + Tlast, source column Lambda_z, AUCINF_pred
2nd worksheet: Sort for the columns as above + Time, source column Predicted
Click on Sort Map, ☑ Use internal Worksheet:
In the top row(s) there should be our usual mappings.
In the last row: Column Worksheet 1 Sorts: Tlast, Worksheet 2 Sorts: Time.- Execute to see the predicted concentrations at Tlast.
Hope that helps.
Prioritising this issue would be nice. C(TAU) is one of the mandatory PK metrics according to the EMA’s MR guideline (11/2014). If the half life is relatively short and there are deviations from scheduled sampling at TAU using the predicted C(TAU) will reduce variability. Helps a lot if the concentration at TAU is missing (like in Dieter’s case). I’m using this approach for years.
Thanks a lot, Helmut! I fully agree to your comment regarding priority
0 user(s) are reading this topic
0 members, 0 guests, 0 anonymous users