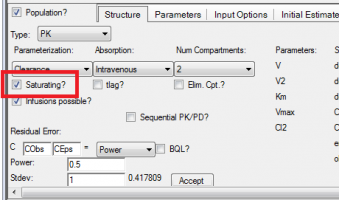
QUESTION: Where on the structure tab did you select the M-M elimination?
ANSWER: Simply check the “Saturating” box on the structure tab to specify M-M elimination.
QUESTION: Can you comment on the error model to use?
ANSWER: One usually starts with the default Additive error model in the first run, and then switches to a different error model to improve the fitting. For example, the Multiplicative error model is useful to help fit smaller concentrations when concentrations span several orders of magnitude. In the particular case of this exercise, when you look at the residuals for an additive error model this shows the need for weighting, but if you then switch to multiplicative it may "over" weight the data. The solution is to use something in-between additive and multiplicative. Since additive corresponds to a power of 0, and multiplicative a power of 1, the solution is to try a power of 0.5. That particular model also has a name - Poisson error.
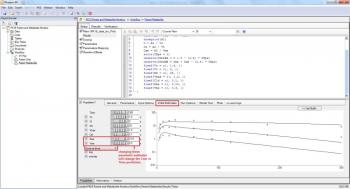
QUESTION: How do you get the initial estimates for Kme and Vme?
ANSWER: You can visualize Cme vs Time. There you can adjust the values to determine initial estimates for Kme and Vme. See example screenshot below.
QUESTION: Is it a simple extension to apply this to multiple subjects? (Adding random effects, etc)
ANSWER: Yes, it is simple to apply this model to multiple subjects. Simply add a subject ID column to the input dataset (with associated observations) and then map the subject ID column as a Sort to get individual subject parameters, or as ID to get one set of PK parameter estimates for all subjects. There is a random effects tab in the Phoenix model object to do PopPK modeling, and this is covered in the NLME training series.
QUESTION: What if there are more than one metabolite, you only measure one metabolite, and you don't know what percentage of the parent dose gets metabolized to that metabolite?
ANSWER: You would need to add an elimination compartment that covers any alternative elimination pathways. There is morediscussion in one of the earlier replies to this topic in the forum.
https://support.cert...inetics/?p=4011
QUESTION: Why is there a Vt in the model if we are not measuring the drug concentration in the tissue/peripheral compartment?
ANSWER: This is a standard situation for a 2-compartment model: we typically don’t have measurements from tissues. We are just fitting our parameters to a biphasical curve of the concentrations of the parent compound.
QUESTION: For the metabolite, A0 is used as the parent drug dose. Should this be adjusted for the molar ratio of the parent and metabolite?
ANSWER: Both measurements, parent and metabolite, are expressed in molar units – not in mass units. That means, molar ratios have already been taken into account.
QUESTION: Regarding QQ plots in the data output: what is the difference between conditionally weighted, individually weighted and weighted residuals?
ANSWER: WRES, IWRES and CWRES are specific model diagnostics. Their calculation is described in:
Hooker, Andrew C., Christine E. Staatz, and Mats O. Karlsson. "Conditional weighted residuals (CWRES): a model diagnostic for the FOCE method."Pharmaceutical research 24.12 (2007): 2187-2197.
QUESTION: Does this work for cassette type dose studies, where we may administer three or more test materials together, all at very low levels? Any obvious upper limitation?
ANSWER: I suppose, you are talking about different parent compounds, not about several parent compounds with their respective metabolites. This would be a different model since I believe the different test materials share elimination pathways so you need to consider drug-drug interactions. Although this might not be the case if levels are really low, in which case you would model each test material on its own.
There are no upper limitations regarding the number of metabolites (see question below)
QUESTION: Are the fixed effects statements the only place to enter initial estimates? It would mean having a different model for each individual if one has a study with multiple individuals (and different initials for each individual)
ANSWER: This has been addressed in an earlier reply on this topic: If you enter initial estimates as part of the PML code or via the tabs Parameters/Fixed Effects you are correct that only a single set of initial estimates can be entered and applied to all subjects. However, if you use the main mapping input, you can enter different values by clicking on the Parameters option (under Setup) and then clicking on rebuild. Or alternatively you can browse for another file (like NCA output) and map them as initial estimates
QUESTION: Is the reason you used the population mode is to just overlay the profiles at different doses? what will happen if you unchecked that?
ANSWER: Correct, if you uncheck population box, the profiles are not overlaid anymore. This was shown in the demo.
QUESTION: What if you would like to model metabolite formation instead of entering fixed Kme and Vme values
ANSWER: In this exercise metabolite formation was modeled through a saturating elimination of the parent compound, here is the corresponding PML statement
deriv(A0 = (Vmax * C / (Km + C)) - (A0*Kme))
the first part describes formation through Michaelis-Menten followed by first order elimination.
QUESTION: Parent will be metabolized to form only one metaboilite. Is that correct?
ANSWER: Correct. If you have other elimination pathways you would need to add another elimination compartment
QUESTION: How many metabolites can you deal with in PHOENIX?
ANSWER: To my knowledge there is no restriction in the number of differential equations (each metablolite would be added as a differential equation statement) to be used in the Phoenix model. The challenge would be to determine initial estimates for all of those.
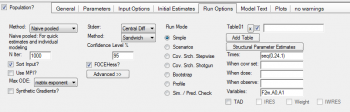
QUESTION: How to estimate the fraction of parent drug converted to metabolite?
ANSWER: Since we have IV infusion data we can assume that 100% of the parent dose is available. In that case you can enter a statement to the PML code that divides the amount of metabolite that is formed by elimination, A0, by the dose:
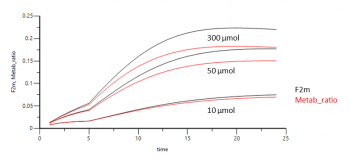
F2m = A0/ A1
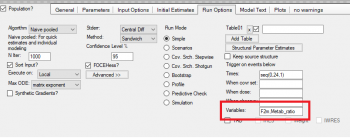
You can report this estimate by adding a Table under the Run Options tab and enter ‘F2m’ as parameter you want to report. For the time you can enter a sequence statement, e.g. seq(0,24,1) that tells the program to report the parameter F2m for the duration of 24 hours with a 1 hour interval:
The time curve of F2m shows a plateu at the end, it converges to the estimate for the fraction metabolized.
QUESTION: If the dosing is not infusion and just single time injection in animals how to set the rate per min?
ANSWER: If there is no infusion, just an IV Bolus dose, there is no need to specify a rate. If you switch off the box for ‘Infusion possible’, the interface would remove the input for dose rate.
QUESTION: Is the metabolite being treated as 1 compartment behavior while the parent is being treated as 2 compartment behavior?
ANSWER: Yes, the concentrations of the parent compound follow a biphasic curve so a 2-compartment model fits nicely. The slope for the metabolite follows first order elimination, so one compartment is suitable.
QUESTION: How easily can we expand on this? For example can we have the parent modeled to 2-comp, the metabolite to MM, and a 2nd metabolite to 1-Comp?
Answer: This has been extensivley discussed in an earlier reply to the topic in the Forum:
https://support.cert...olite-kinetics/
QUESTION: Why can’t you use graphical model for this parent -metabolite kinetics?
ANSWER: You can use the graphical model builder for this as well.
QUESTION: To find the average concentration of 3 dose (example), does it necessary to mix whole concentration?
ANSWER: From your question it is not clear what you want to achieve. Can you please describe the problem in more detail. If possible, you may want to share your dataset of project file.