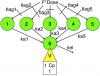
For a modified release drug formulation deconvolution is accomplished within WinNonLin by the deconvolution tool. Another optional way of looking at this is to consider the model whereby an oral dose of drug (D mg) is released into the gastrointestinal tract with release rate R. The drug is then absorbed with absorption rate K01 into the central compartment, the transfer rate into the peripheral compartment is K23; transfer back is K32. The elimination rate from the central compartment is K10.
From plasma concentrations following oral dose of an immediate release formulation I have estimates of K10, K23, K32 and I have the plasma concentrations of MR formulation following oral dose. I want to deconvolute the MR plasma concentrations to obtain the the invivo rate release profile (R values), which are the invivo release rates into the GI tract (not the invivo absorption rate).
Here is some embryonic code and I am looking for suggestions as to making this code work in WinNonlin or Phoenix. For example how to get the dose included in the model and get F in units of the concentration. So I I have NCON at 3 by that I mean to have the single dose defined and the time (0) and amount of the dose.
NFUNCTIONS 1
NDERIVATIVES 2
NPARAMATERS
NCON 3
PNAMES 'R' 'K10', 'K01', 'K23', 'K32', 'V_F'
START
Z(1) =0
Z (2) =0
Z(3) = 0
END
DIFFERENTIAL
D(Z1)= R - k01 *Z(1)..................................................Gastrointestinal tract
D(Z2) =k01*Z(1)-(k10 + k23)*Z(2).............................Compartment 2 {central}
D (Z3) =k23* Z(2) -k32*Z(3).......................................Compartment 3 {Peripheral}
END
FUNCTION 1
F = Z(2)/V_F
END
EOM