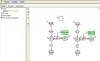
Thought I would share this with the forum since I ended up working a way to start with the graphical interface, then flip over to edit as textual for this one (Thanks again to Jason C from Pharsight who has been my guide as I fumble though becoming a pharmacometrician). I think its worth considering this approach for anyone looking to simultaneously model two formulations from a crossover design where the two formulations will share the same estimates for Clearance and Volume and individually fit for ka. In addition, by setting a reference formulation with F=1, you can then fit the relative F for the test product this way.
In this case, first we started with the graphical interface for a standard 2-compartment model parameterized for clearance and extravascular administration. The input file needs to contain separate files for each formulation for dose and Cobs and mapped accordingly.
For whatever reason I can't copy the image into this file from the Phoenix model object to demonstrate how to start in the graphical mode, but below is the closed form text.
Elliot
test(){
deriv(A1 = - (Cl * C) + (Aa * Ka)- (Cl2 * (C - C2)))
urinecpt(A0 = (Cl * C))
deriv(Aa = - (Aa * Ka))
deriv(A2 = (Cl2 * (C - C2)))
deriv(A3 = (Aa2 * Ka2)- (Cl2 * (C3 - C4))- (Cl * C3))
deriv(Aa2 = - (Aa2 * Ka2))
deriv(A4 = (Cl2 * (C3 - C4)))
urinecpt(A0ref = (Cl * C3))
C = A1 / V
dosepoint(Aa, bioavail = (F), idosevar = AaDose, infdosevar = AaInfDose, infratevar = AaInfRate)
C2 = A2 / V2
error(CEps = 0.388647)
observe(CObs1 = C * (1 + CEps))
C3 = A3 / V
dosepoint(Aa2, bioavail = (1), idosevar = Aa2Dose, infdosevar = Aa2InfDose, infratevar = Aa2InfRate)
C4 = A4 / V2
observe(CObs2 = C3 * (1 + CEps))
stparm(V = tvV * exp(nV))
stparm(Cl = tvCl * exp(nCl))
stparm(Ka = tvKa * exp(nKa))
stparm(V2 = tvV2 * exp(nV2))
stparm(Cl2 = tvCl2 * exp(nCl2))
stparm(F = tvF * exp(nF))
stparm(Ka2 = tvKa2 * exp(nKa2))
fixef(tvV = c(, 202053, ))
fixef(tvCl = c(, 55033.6, ))
fixef(tvKa = c(, 2.67803, ))
fixef(tvV2 = c(, 1052530, ))
fixef(tvCl2 = c(, 95295.7, ))
fixef(tvF = c(, 0.879595, ))
fixef(tvKa2 = c(, 0.709612, ))
ranef(diag(nV, nCl, nKa, nV2, nCl2, nF, nKa2) = c(0.021506572, 0.23185064, 0.072966132, 0.022392015, 0.15125611, 0.076635195, 0.073511574))
}