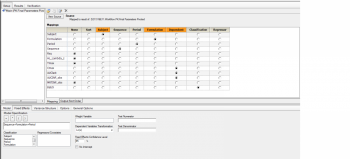
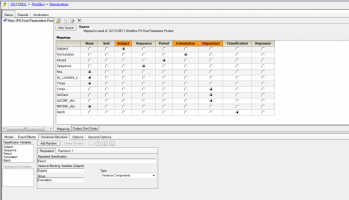
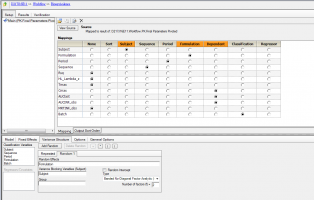
We conducted a replicate study which is RRT, RTR and TRR. We completed 90%CI with default setting, such as attachments. However, the study was completed with 5 groups, so that I want to know how group effect is analyzed. I have searched BABE forum for couple of days. but still can not get the answer. Would anyone please guide me how to do it in the WLN? thank you.
I have seen a paper use: Group+Sequence+Period+Treatment+Random errors where Random errors is Random within and between subject errors. However, what about Formulation*Group such in the 2x2 crossover study? I am not so sure it is correct.
Charlie
Edited by tsang0323, 13 April 2016 - 01:46 AM.