Hi mouksassi,
if you can share the project I can try to help more
I will ask my supervisor to see if I can share the data. He is out of town.
1) My purpose is to use my current SAD data to simulate the MAD regimen in new subjects. So that means I need use pop model as you mentioned below?
SM: Yes.
2) After I ran the pop model, I only found a set of PK parameters in theta. Can we get individual parameters from pop model?
SM: Yes ask for the table and don't forget to ask for the parameters. If you are using a Race categorical effect you should get an StrCovCat sheet. This will have the individual parameters values as well.
A Race categorical effect means that when we add a covariate we set its type as categorical when we run a pop model?
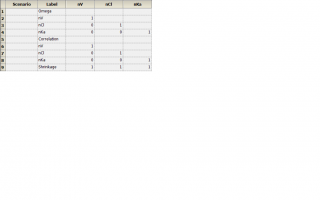
In my pop model with race as a covariate, I did find individual parameter in StrCovCat but the parameters for each subject are the same. I think that is because I used naive pooled engine and eta=0 (please see the attachment named “omega”).
3) Another confusing issue is omega. I always get 1 in the omega even I tried to run the example in Phoenix NLME 1.3 User's Guide. Some problem with my software?
You might not be using the population engine or something not mapped correctly.
We need to see the project to help.
I used Naive pooled engine. In the NLME guide, it is said that "All ETAs are forced to zero, and no OMEGA parameters are computed - only THETA and SIGMA" when using this engine. I think that is the reason. Naïve pooled engine is the only method in my software. Do other methods need extra license? If I only have Naïve Pooled engie, dose this mean that I can not run pop model?
4) In the Phoenix NLME 1.3 User's Guide (page 23), it is said that select the Closed form have some disadvantages and one is that the differential system being converted must be linear (no nonlinear kinetics). Dose this mean that if we deselect the Close form we can pool all data no matter they are linear PK or nonlinear PK?
Hi closed form only exist for some of the differential equation models so these are not related. As I mentioned you can use DOSE as a covariate on CL but in reality it is not a dose effect it is a receptor being saturated due to high concentration or something else the alternative to using DOSE would be to use a Michaelis Menten model, TMDD, non linear binding etc.
if you select saturating then your closed form solution disappear the software take care of the rest.
Pooling all data is a user decision ideally you want to fit all data together so you integrate all the knowledge. The challenge is to come up with a model and theory that fit ALL the data.
If the model can not fit all data well can I split the pooled data into 2 parts (one include the data of 0.3-2mg, and the other includes the data of 4 and 8 mg) and then analyze them seperately? Do you have any other idea?
5) That is exactly what I want to do!
When I use pop model to simulate what file should be mapped to the Main in Setup?
Just a file with the dosing regimen you want you can list all doses but you can also use the ADDL feature which is handy if you want regular dosing intervals. If there is covariate in your model you have to chose a covariate value that you want to simulate for
ID TIME DOSE ADDL II RACE
1 0 1 2 24 2
equivalent to :
ID TIME DOSE RACE
1 0 1 2
1 24 1 2
1 48 1 2
6) My estimate of dVdRACE2 and dCldRACE2 are negative. And most CV%s are more than 30%. I tried to use 2 compartment model but it didn't improve the result. What can I do?
I need to see how you parametrized you model to be able to comment. how many subjects you have ?
I totally have 36 subjects.
7) So in the Parameters I can use the pop parameter?
Yes and then you will be randomly sampling subjects based on the omega you estimated but first we need to fix why your omega are 1
I am a little confused. Do you mean I need to input some values in the Ramdom Effect when I run the individual or pop simulation? These values are the values of nV, nCl etc from the Omega?
8) Is the distribution of covariate information a must in my regimen? Now I just want to simulate different dose regimens and the new subjects will be enrolled with the similar criterion.
Can I input different MAD schedule like this:
see above and it is up to you if you have other than race covariate effects you need to consider it think whether you want to simulate what happens for a 70 kg RACE =2 subject or whether you want to simulate a realsitic distribution of covariate including correlated weight, race,sex ( best distribution you can use if you have tons of data is your own data !)
ID TIME DOSE ADDL II RACE
1 0 1 13 168 2
2 0 1 13 168 2
3 0 1 13 168 2
4 0 1 13 168 2
5 0 1.7 0 0 2
5 168 1 12 168 2
For “simulate a realsitic distribution of covariate including correlated weight, race,sex”, I am confused. Could you please explain it with more detail?
Furthermore, the example listed above is very helpful and I did the simulation accordig to this. Below are the steps and the results.
The regimen format looks like this
ID TIME DOSE Dose
1 0 1 1000
1 24 1 1000
1 48 1 1000
2 0 1.7 1700
2 24 1 1000
2 48 1 1000
3 0 0.5 500
3 24 0.5 500
3 48 0.5 500
Please see the attachment “regimen” for more detail. I only list ID time and dose because I only want to see the MAD concentration in subjects with similar covariates.
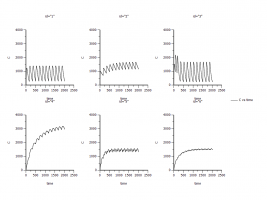
1) First, I simulated the MAD with individual model.
Map the sort, Aa (Dose in ng), and Time in Mappings.
Use the pop paramters (I ran two pop models, one is without covarites and the other is with covarites. Which set of pop parameter should I use? Here I use the pop parameter from the model without covarites).
Ramdom Effects: I didn’t input anything because I don’t know what to use.
Run options: #=1000, Max X=2016, and Y=C
The Residual error is mixed ratio
Run the model
It works! But I don’t know whether the results are correct since I didn’t use the ramdom effects. Please see the attachment “Ind simulation”.
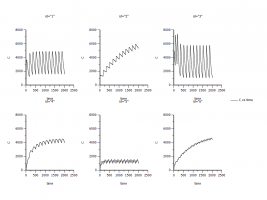
2) Then I smimualted the MAD with pop model.
Copy and paste the individual model.
Check pop.
Change to ID=regimen and keep the remains.
N Iter=0 (actually I don’t know why I need do this. I saw this at other people’s post)
Run options: Main: #=1, X axis=t, Pred.=None; Add sim table: Times=seq(0,2016,1), variables=C
Run the model
The same doubt: the results are correct since I didn’t use the ramdom effects? Please see the attachment “XY plot pop simulation #1 none pred”.
3) Base on the mode 2) I changed something in Main in Run options. I set Pred.=additive and checked the Pred. Variance. Corr. Checkbox and run the model. The result of 3 is different from the result of 2 (Please see the attachment “XY plot pop simulation additive pred”).
The pred. is only for preditive? For simulaiton, we don’t need pred.? So set pred.=None (model2)) is more reasonable?
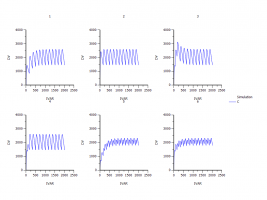
I compared the plots of 1) , 2) and 3). They are different. Which result I should use?
I also fitted all the data with PK libaray model (here I used uniform as weighting). I summarized the individual PK parameters and use the mean PK parameters (the CV% is huge) to simulate the MAD with PK libaray model.
WinNonlin is so powerful and provides so many choice so I am lost J Which one is better?
Thank you for your time again!
LLLi
Edited by LLLi, 29 July 2016 - 07:43 PM.